Image data used in this challenge were acquired on a 3T scanner at the UMC Utrecht (the Netherlands). For each of the 30 subjects, fully annotated multi-sequence (T1-weighted, T1-weighted inversion recovery and T2-FLAIR) scans are available. The 30 subjects include patients with diabetes, dementia and Alzheimers, and matched controls (with increased cardiovascular risk) with varying degrees of atrophy and white matter lesions (age > 50).
Details
For each patient, the following sequences are provided:
| Sequence | Details | TR (ms) | TE (ms) | TI (ms) |
|---|---|---|---|---|
| T1 | 3D T1-weighted sequence | 7.9 | 4.5 | |
| T1-IR | Multi-slice T1-weighted inversion recovery sequence | 4416 | 15 | 400 |
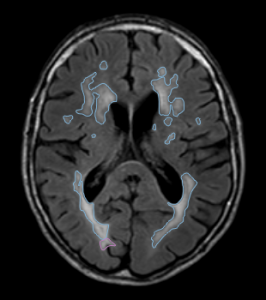
| T2-FLAIR | Multi-slice T2 FLAIR sequence | 11000 | 125 | 2800 |
All scans have a voxel size of 0.958mm × 0.958mm × 3.0mm and all are aligned. The scans are bias field corrected using the N4ITK algorithm, but the original data is also available.
Some of the T1-IR images contain artifacts at the bottom of the scan. These artifacts occur often in clinical scans, and it is therefore interesting to know how automatic segmentation methods that use these scans perform under these circumstances.
Manual reference standard
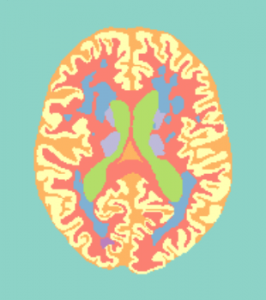
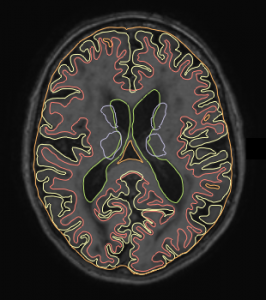
Training data (7 subjects) will be accompanied with a file containing the manual reference standard, containing the following 11 labels:
| Label | Description |
|---|---|
| 0 | Background |
| 1 | Cortical gray matter |
| 2 | Basal ganglia |
| 3 | White matter |
| 4 | White matter lesions |
| 5 | Cerebrospinal fluid in the extracerebral space |
| 6 | Ventricles |
| 7 | Cerebellum |
| 8 | Brain stem |
| 9 | Infarction |
| 10 | Other |
The objective of this challenge is to automatically segment labels 1 to 8. Ground-truth labels 9 and 10, i.e. infarctions and ‘other’ lesions, will be excluded in the evaluation. This means that those voxels will have no effect on the scores; you may label these voxels as gray matter, white matter, or any other label. The background (label 0) will not be treated as an ‘object’ itself, but any false-positive label (1 to 8) in the background will negatively influence the scores.
Algorithms that only segment gray matter, white matter and cerebrospinal fluid should merge labels 1 and 2, 3 and 4, and 5 and 6, and label the output as either 0 (background), 1 (gray matter), 2 (white matter) and 3 (CSF). The cerebellum and brain stem (label 7 and 8) will in that case be excluded from the evaluation. The results on the merged labels will be ranked separately.
Notes on the manual segmentations
- White matter lesions were segmented on the FLAIR scan.
- The outer border of the CSF was segmented using both the T1-weighted scan and the T1-weighted inversion recovery scan.
- All other structures were segmented on the T1-weighted scan (0.958mm × 0.958mm × 3.0mm).
- Vessels were not segmented separately. The CSF segmentation therefore also includes the superior sagittal sinus and transverse sinuses.
- The cerebral falx is also included in the CSF segmentation.
- Segmentation labels
- T1
- T1-IR
- T2-FLAIR
Additional data
Participants are allowed to include publicly available data to enrich the training set. This must be mentioned in the description that is submitted with the method. New manual annotations on public data should be shared.